Cutting out the amplicon with restriction endonuclease – a way to exclude the false results of RT PCR with TaqMan probe
Akishev A.G., Netesova N.A., Abdurashitov M.A., Degtyarev S.Kh.
1 SibEnzyme, Novosibirsk
2 Epigenlab, Novosibirsk
* corresponding author: Alexander Akishev, SibEnzyme, 2/12 Ak.Timakov Str., Novosibirsk 630117, Russia; Tel: +7 383 3334991; Fax: +7 383 3336853; E-mail: aki@sibenzyme.ru
Real-time PCR with TaqMan probe (RT PCR) is one of the main methods for analyzing the structure of human DNA and is widely used in medicine. In this work, PCR RT of 4 fragments of human genomic DNA (chr1: 90717178-90717300, chr9: 97464604-97464756, chr11: 65647215-65647307, chr17: 4560027-4560175) was performed with DNA preparations from human blood obtained both by the standard method of phenol deproteinization and isolated on columns. Preparations of the original DNA and DNA after a treatment with a restriction endonuclease that cuts out the amplified fragment were used in PCR. In the case of native DNA preparations purified by column, PCR of 3 from 4 DNA fragments provided overestimated Cq values. In the case of the phenol purification, overestimated Cq values for native DNA preparations were obtained for 2 from 4 DNA fragments. For DNA preparations obtained by the column purification, treatment of DNA with a restriction endonuclease that cuts out the amplified fragment leads to normal Cq values for all four analyzed DNA fragments. Thus, RT PCR of native DNA obtained by the column purification may result in an overestimation of Cq, which is avoided if DNA previously has been hydrolyzed by a restriction endonuclease that cuts out the amplicon.
Key words: restriction endonucleses, DNA preparations from human blood, RT PCR
DOI: 10.26213/SE.2021.45.29.001
Citation:
Akishev A.G., Netesova N.A., Abdurashitov M.A., Degtyarev S.Kh. (2021) Cutting out the amplicon with restriction endonuclease – a way to exclude the false results of RT PCR with TaqMan probe, DNA processing enzymes, vol 2021(1), DOI: 10.26213/SE.2021.45.29.001
This work is available at Creative Commons licence «Attribution-NonCommercial» 4.0 International.
INTRODUCTION
Real-time PCR with TaqMan probe (RT PCR) is widely used in the analysis of the human DNA structure and is one of the main diagnostic methods in medicine. Earlier, we showed that a preliminary hydrolysis of DNA with restriction endonuclease that cuts out the amplified fragment allows to conduct comparative PCR studies of many DNA preparations [1-4]. In this work, we performed RT PCR study of 4 fragments of human genome in DNA preparations from human blood obtained both by the standard method of phenol deproteinization and purified by the column. PCR was performed both on the original DNA and after its hydrolysis with restriction endonuclease which cuts out the amplified fragment.
MATERIALS AND METHODS
Materials. Phenol (JSC “Reactiv”, Novosibirsk), chloroform (JSC “Reactiv”, Novosibirsk), isoamyl alcohol (JSC “Reactiv”, Novosibirsk), RNAse A (LLC “Samson-Med”, St. Petersburg), SDS (LLC “Helicon”, Moscow) were used in the work. “PCR kit with DNA pretreatment”, “kit for DNA purification from a light fraction of the blood cells”, oligonucleotides, Bst2UI, HinfI, MspI restriction endonucleases, bidistilled water and buffer solutions produced by SibEnzyme Ltd.
The material for the study was DNA samples isolated from blood cells of 16 donors. DNA preparations of 8 donors under the numbers 205, 1086, 1087, 1137, 1151, 1205, 1437 and 1487 were obtained by the phenol-chloroform method as described earlier [5] with some modifications. The obtained DNA preparations were treated only with RNase A without an addition of TaqI restriction endonuclease. In the case of other 8 donors under the numbers 232, 235, 582, 558, 559, 604, 605 and 640 DNA was isolated from a light fraction of the blood cells as described earlier [4]. The concentration of the obtained DNA preparations was determined by spectrophotometer.
The restriction mixture was prepared according to the protocol [6]. 250 µl of the restriction mixture from “PCR Kit with DNA pretreatment” were mixed with 1 µl (20 units) of HinfI restriction endonuclease (mixture A) for hydrolysis of DNA fragment 9-604. 250 µl of the restriction mixture from “PCR Kit with DNA pretreatment” were mixed with 1 µl (20 units) of Bst2UI restriction endonuclease (mixture B) for hydrolysis of DNA fragment 1-178. 250 µl of the restriction mixture from of “PCR Kit with DNA pretreatment” were mixed with 1 µl (20 units) of MspI restriction endonuclease (mixture C) for hydrolysis of DNA fragment 17-027 and 11-215.
Preparation of a PCR mixture. 77 µl of a mixture of primers and a probe (10 µM each) specific to the studied DNA fragment and 12.8 µl of Hot Start Taq DNA polymerase (5 units of act/µl) were added to 940 µl of the PCR-mix from “PCR Kit with DNA pretreatment” and mixed.
Hydrolysis of the blood DNA with restriction endonucleases was carried out in test tubes for PCR (200 µl). In each test tube 13.5 µl of H2O and 1.5 µl of DNA with concentration about 18 ng/µl were mixed. 2 tubes with DNA were prepared for each sample. 15.1 ml of restriction mixture was added to one tube. 15.1 ml of mixture A was added to the other tube in the case of analysis of DNA fragment 9-604, 15.1 ml of mixture B was added to the other tube in the case of analysis of DNA fragment 1-178, and 15.1 ml of mixture C was added to the other tube in the case of analysis of DNA fragments 17-027 or 11-215. For the DNA fragment 1-178, incubation was carried out at 60 C, in the other cases – at 37 C. The incubation time was 30 minutes.
PCR. After incubation and a shot centrifugation to collect drops, 30.2 µl of PCR mixture was added to each tube. After mixing, the content of each test tube (60 ml) were dispensed (20 ml each) into the wells of a PCR plate for 96 wells.
To calculate specific primers and probes, we used nucleotide sequences from the GenBank database (http://ncbi.nlm.nih.gov/genbank) according to the version of the human genome GRCh38/hg38, the set of programs “Vector NTI 11.5 “(Invitrogen, USA) and the online resource “BLAST” (http://blast.ncbi.nlm.nih.gov). The structure of the primers and the fluorescently labeled probe used in the work is shown below:
9-604z 5′ FAM-TGTGAGCCACTGTGACCAGCCCCC-BHQ1 3′(24)
9-604d 5′ CCTCGGCCTCCCAGAGTGC 3′(19)
9-604r 5′ CCTGCTGCCATCTTTGGTTCC 3′ (21)
17-027z 5′ TCAAAGGTCCCCAGCCCCCAA 3′(21)
17-027r 5′ CCAAGCCAGCCTCTGTCCCA 3′(20)
17-027d 5′ GGGGTGTGGCAGGCAGGAG 3′(19)
11-215z 5′ CCACCGCCCGAGGGATGAAGACC 3′(23)
11-215d 5′ TGGTGCCCTCGGGTAAGCG 3′(19)
11-215r 5′ CCC AGG CCA CAC GGA ACT TTC 3′(21)
1-178z 5′ CCGCCGTTCAGCAGCCGCCC 3′(20)
1-178r 5′ CGGCGGCAACATTGTTTGGT 3′(20)
1-178d 5′ GGC CCC TTC CAT TGT CAT TGC T 3′(22)
RT PCR was performed according to the manufacturer’s Protocol [7] in a volume of 20 µl in the thermocycler CXF-96 (“Bio-Rad”, USA) according to the following program:
for fragments 9-604, 17-027, 1-178 :
- at 95°C 3 min;
- 5 “blind” cycles: 95°C – 10 sec, 69°C (with a decrease of 0.7°C in each subsequent cycle) – 20 sec, 72°C – 10 sec, 75°C – 5 sec;
- 40 cycles: 95°C – 10 sec, 65°C – 20 sec (with detection of the fluorescent signal in the FAM channel), 72°C – 10 sec, 75°C – 5 sec;
for fragment 11-215:
- at 95°C 3 min;
- 5 “blind” cycles: 95°C – 10 sec, 69°C (with a decrease of 0.5°C in each subsequent cycle) – 20 sec, 72°C – 10 sec, 75°C – 5 sec;
- 40 cycles: 95°C – 10 sec, 66°C – 20 sec (with detection of a fluorescent signal in the FAM channel), 72°C – 10 seconds, 75°C-5 sec.
Upon completion of PCR an average value of Cq and a value of the standard deviation for the analyzed samples were determined using the software of the amplifier “Bio-Rad CFX Manager v. 2.1”. The data obtained were entered in the table.
RESULTS AND DISCUSSION
In this work, we studied DNA preparations from the blood of 16 donors obtained both by the standard method of phenol purification and using a DNA isolation kit [8]. Among the donors there were 7 men and 9 women. Table 1 shows the gender and age of donors (columns 2 and 3, respectively), as well as the concentration of the obtained DNA preparations (column 4) and the characteristics of their purity, determined by the ratio of optical density at 260 nm and 280 nm (column 5). The numbers of DNA preparations obtained by phenol purification are highlighted in blue background.
Table 1.
Gender and age of blood donors, concentration and photometric purity of the obtained DNA.
|
N |
Sex |
Age |
DNA concentration, ng/µl |
А260/А280 |
|
232 |
f |
42 |
74 |
1,80 |
|
235 |
m |
54 |
35 |
1,90 |
|
558 |
f |
68 |
54 |
1,90 |
|
559 |
f |
66 |
20 |
1,90 |
|
582 |
m |
33 |
54 |
1,90 |
|
604 |
f |
36 |
52 |
1.90 |
|
605 |
m |
54 |
50 |
2,00 |
|
640 |
m |
43 |
48 |
1,80 |
|
205 |
f |
72 |
461 |
1.80 |
|
1086 |
m |
36 |
193 |
1.70 |
|
1087 |
f |
51 |
95 |
1.70 |
|
1137 |
f |
59 |
299 |
1.60 |
|
1151 |
f |
50 |
225 |
1.70 |
|
1205 |
m |
65 |
420 |
1.70 |
|
1437 |
m |
68 |
213 |
1.80 |
|
1487 |
f |
22 |
406 |
1.70 |
As can be seen from the table, the isolated DNA preparations have a concentration in the range from 20 ng/ml to 461 ng/ml and a purity of at least 1.6 (the ratio A260/A280).
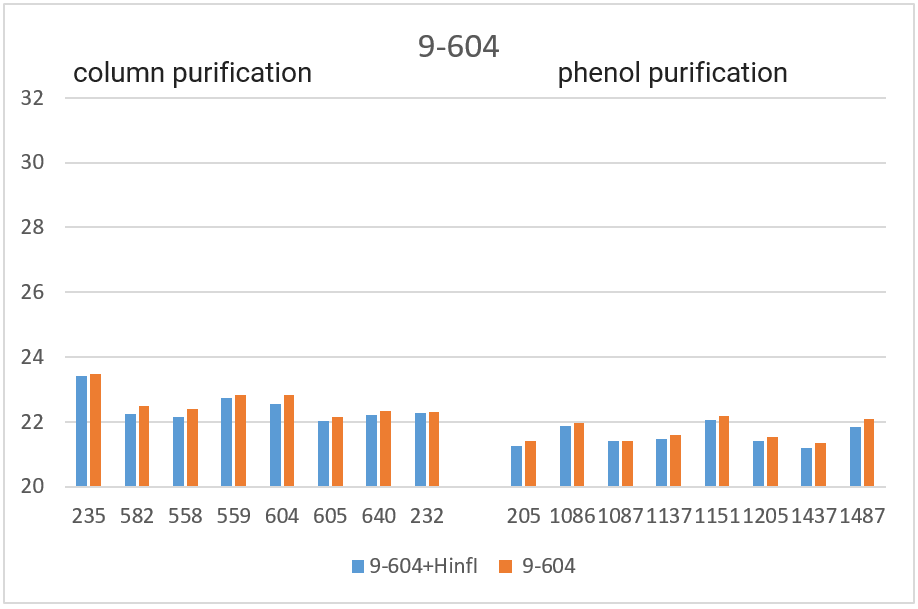
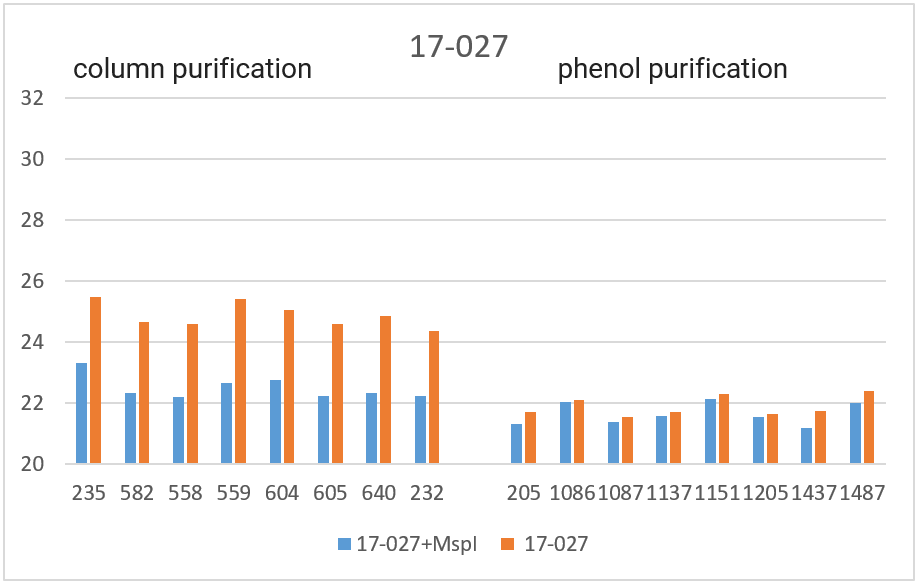
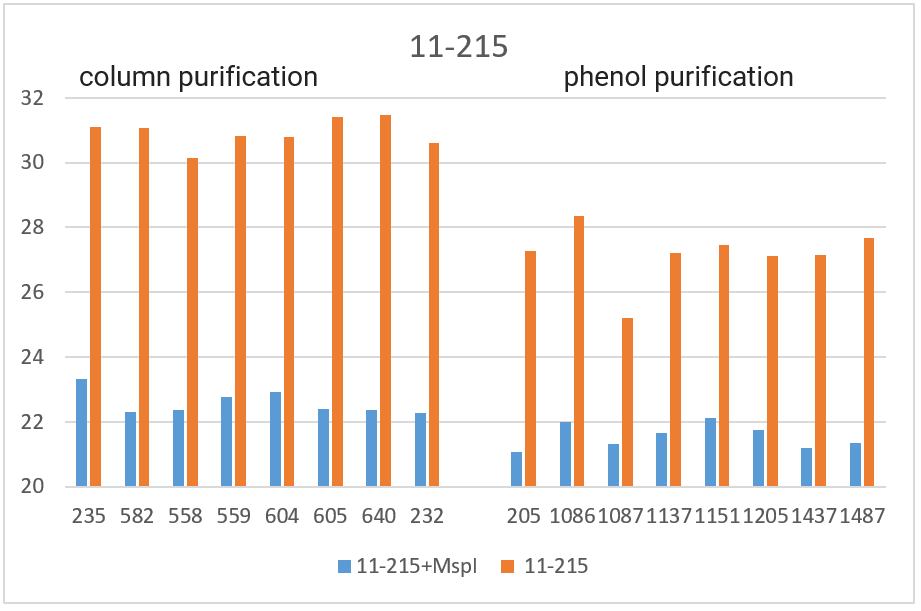
RT PCR was performed with the obtained DNA preparations, both native and after cleavage with one of the three restriction endonucleases (Bst2UI, HinfI, MspI), as indicated in the “Materials and Methods”. For each of the 4 amplified fragments, hydrolysis of DNA preparations with restriction endonuclease Bst2UI (fragment 1-178), HinfI (fragment 9-604) and MspI (fragments 17-027 and 11-215) leads to cutting out the amplified fragment and allows to avoid the influence of the rest of DNA on the PCR results.
We performed RT PCR of 4 DNA fragments, the name of which indicates the position of the 5’-end of amplicon in the genome and includes the chromosome number and the last three digits of its location. For example, the name of fragment 9-604 corresponds to location of its 5’-end at chr9: 97464604.
Figure 1 shows the nucleotide sequence of 4 DNA fragments of genome analyzed in this work. In each DNA fragments, the recognition sites of restriction endonuclease bordering the amplicon are highlighted with a purple background. The binding zones of the forward and reverse primers, as well as the fluorescently labeled TaqMan probe, are highlighted in yellow, blue and green backgrounds, respectively.
Figure 1
The nucleotide sequence of 4 fragments of human genome studied by RT PCR
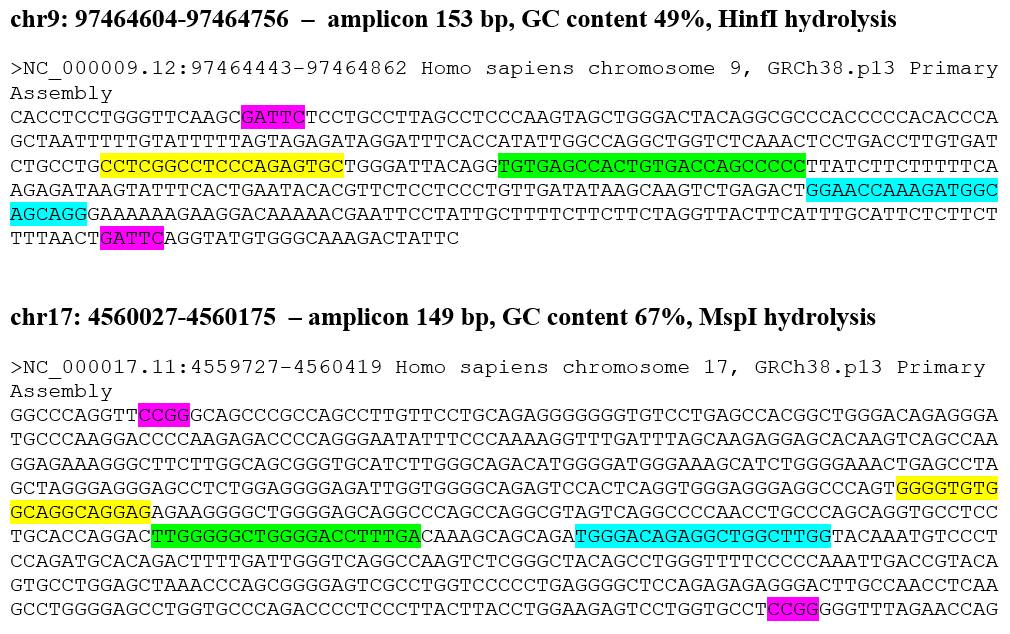

Figure 2 shows the fluorescence curves of RT PCR for DNA samples No. 1151 (phenol purification) and No. 559 (column purification).
Figure 2.
Fluorescence curves of RT PCR of 4 fragments of the human genome in DNA preparations from the donor blood No. 559 and No. 1151.
9-604 |
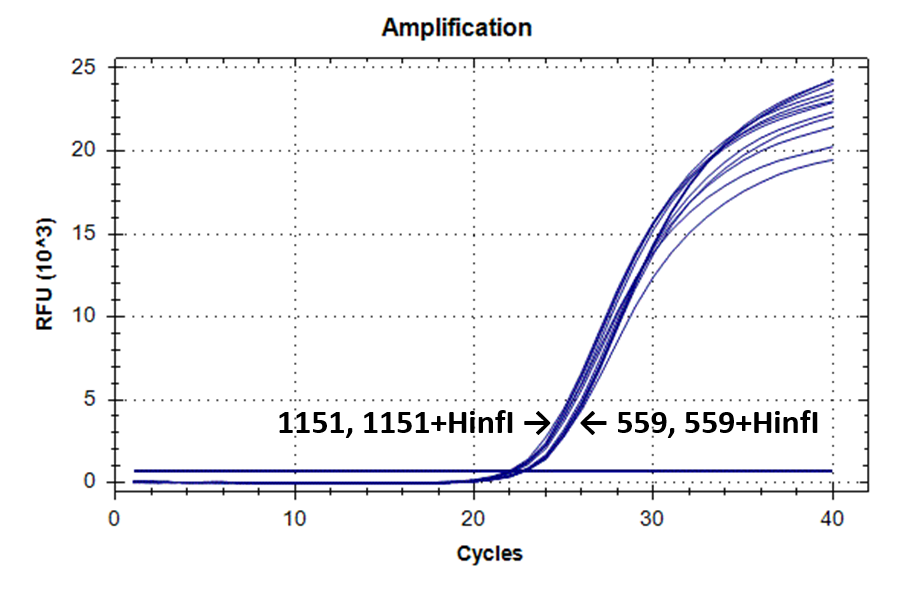 |
17-027 |
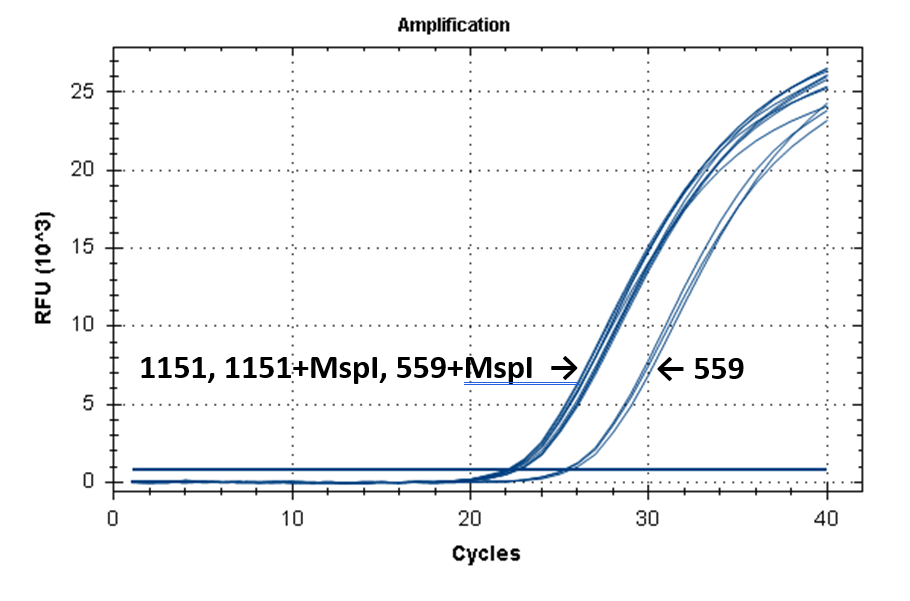 |
11-215 |
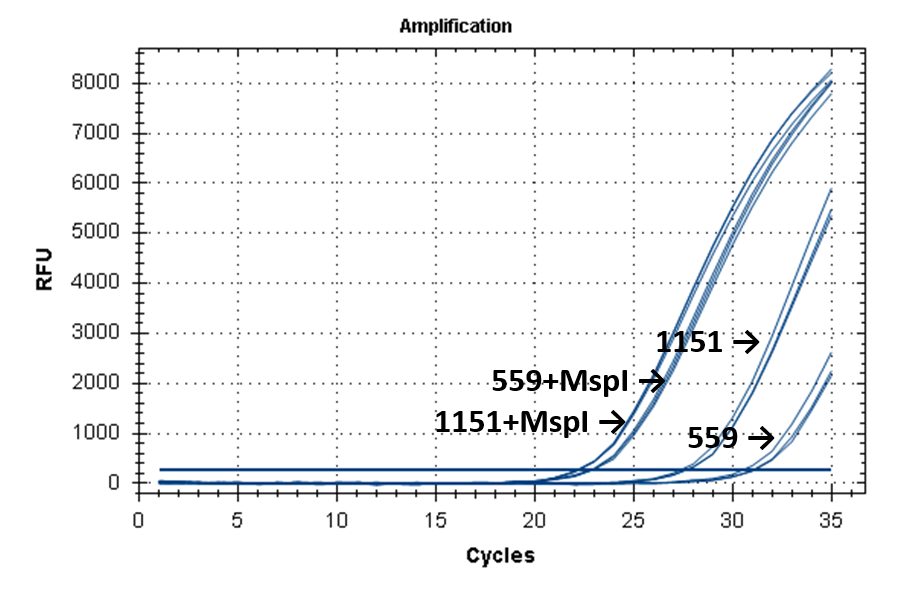 |
1-178 |
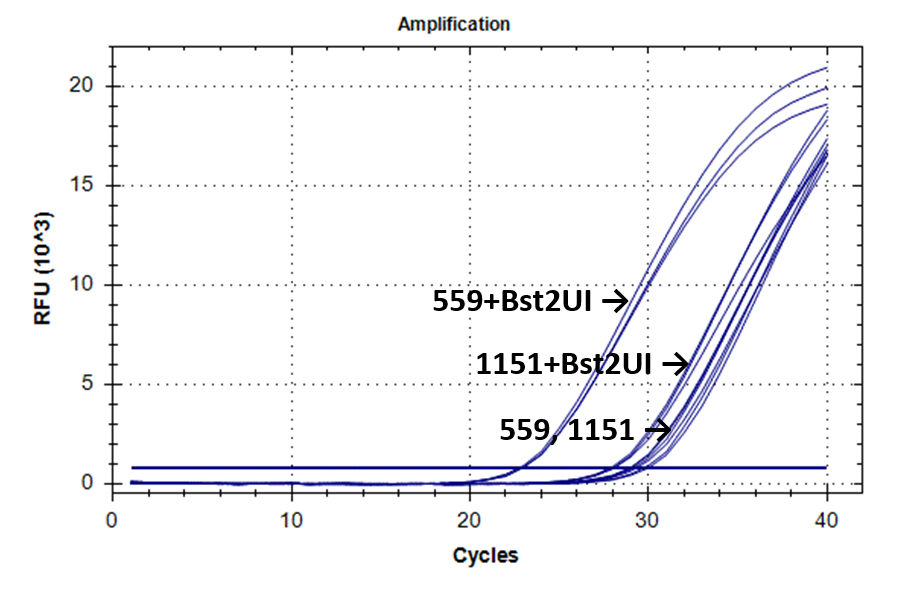 |
Table 2 shows the results of RT PCR of 4 fragments of the human genome in 16 DNA samples from the donor blood.
Table 2.
Cq values obtained by RT PCR of 4 selected fragments of the human genome in 16 the donor blood DNA preparations. The numbers of DNA preparations obtained by phenol purification are highlighted in blue background.
|
№ |
9-604 + HinfI |
9-604 |
17-027 + MspI
|
17-027 |
11-215 + MspI |
11-215 |
1-178 + Bst2UI |
1-178 |
|
232 |
22,27±0,05 |
22,30±0,09 |
22,24±0,09 |
24,35±0,23 |
22,26±0,10 |
30,60±0,53 |
22,34±0,02 |
28,61±0,04 |
|
235 |
23,40±0,07 |
23,47±0,05 |
23,30±0,06 |
25,47±0,17 |
23,32±0,10 |
31,11±0,31 |
23,46±0,06 |
29,51±1,05 |
|
558 |
22,14±0,11 |
22,39±0,06 |
22,19±0,05 |
24,59±0,61 |
22,35±0,05 |
30,13±0,11 |
22,38±0,13 |
28,84±0,43 |
|
559 |
22,72±0,10 |
22,82±0,09 |
22,66±0,08 |
25,41±0,20 |
22,77±0,08 |
30,83±0,34 |
22,79±0,11 |
29,26±0,37 |
|
582 |
22,25±0,04 |
22,49±0,08 |
22,31±0,06 |
24,66±0,13 |
22,31±0,11 |
31,08±1,12 |
22,30±0,07 |
28,63±0,16 |
|
604 |
22,54±0,03 |
22,84±0,03 |
22,76±0,21 |
25,05±0,28 |
22,91±0,28 |
30,79±1,36 |
22,58±0,03 |
28,86±0,09 |
|
605 |
22,02±0,06 |
22,14±0,06 |
22,23±0,11 |
24,60±0,06 |
22,41±0,13 |
31,41±1,28 |
22,10±0,01 |
29,07±0,23 |
|
640 |
22,22±0,07 |
22,34±0,06 |
22,34±0,03 |
24,86±0,09 |
22,36±0,07 |
31,47±1,13 |
22,21±0,01 |
28,90±0,33 |
|
205 |
21,26±0,05 |
21,41±0,07 |
21,27±0,05 |
21,30±0,12 |
21,06±0,04 |
27,27±0,31 |
23,87±0,07 |
26,36±0,21 |
|
1086 |
21,87±0,05 |
21,96±0,06 |
22,02±0,17 |
22,11±0,06 |
21,99±0,09 |
28,35±0,38 |
27,94±0,27 |
28,99±0,17 |
|
1087 |
21,42±0,10 |
21,41±0,07 |
21,39±0,04 |
21,55±0,03 |
21,32±0,04 |
25,21±0,03 |
28,45±0,12 |
28,81±0,31 |
|
1137 |
21,46±0,06 |
21,60±0,18 |
21,56±0,06 |
21,70±0,10 |
21,65±0,10 |
27,21±0,11 |
27,36±0,37 |
28,99±0,17 |
|
1151 |
22,05±0,08 |
22,19±0,11 |
22,13±0,04 |
22,28±0,07 |
22,11±0,04 |
27,47±0,16 |
27,92±0,13 |
29,30±0,54 |
|
1205 |
21,41±0,09 |
21,54±0,08 |
21,54±0,09 |
21,65±0,08 |
21,74±0,28 |
27,13±0,13 |
26,71±0,29 |
28,00±0,16 |
|
1437 |
21,20±0,06 |
21,34±0,10 |
21,18±0,01 |
21,75±0,05 |
21,20±0,18 |
27,16±0,23 |
23,02±0,05 |
25,66±0,14 |
|
1487 |
21,84±0,06 |
22,09±0,06 |
22,00±0,02 |
22,39±0,06 |
21,34±0,15 |
27,66±0,28 |
24,66±0,22 |
27,04±0,35 |
As can be seen from Table 2, for each DNA sample, the Cq values obtained by RT PCR on native DNA are either higher or coincide with the Cq values obtained by RT PCR on hydrolyzed DNA The Cq values for all 16 samples of the native DNA in the case of fragment 9-604 (column 3) and for 8 preparations of the native DNA obtained by phenol purification in the case of fragment 17-027 (the lower half of column 5), as well as the Cq values for hydrolyzed DNA samples in the case of all 4 fragments (with the exception of fragment 1-178 on DNA preparations obtained by phenol purification, columns 2, 4, 6 and the upper half of column 8) practically coincide for each of the DNA samples. These similar Cq values for each DNA sample (the basic value) probably correspond to the real Cq value associated inversely with the number of analyzed molecules of human genomic DNA, in which each of the four fragments is represented in one copy.
The higher Cq values are observed for three fragments. 1) in the case of fragment 17-027, the Cq values for native DNA preparations after the column purification are 2 – 2.5 cycles higher than the basic values (the upper half of column 5); 2) in the case of fragment 11-215, the Cq values are higher for about 6 cycles for DNA preparations after phenol purification, and about 8 cycles for DNA preparations after column purification (column 7); and, finally, 3) in the case of fragment 1-178, the higher Cq values are observed not only in the case of all native DNA preparations (column 9), but also in the case of DNA preparations after phenol purification, which were hydrolyzed by Bst2UI restriction enzyme (the lower half of column 8).
In Figure 3, the obtained Cq values are presented in the form of diagrams. As can be seen from the diagrams, after DNA digestion with a restriction enzyme that cleaves out the amplified fragment, the Cq values for all fragments (except 1-178 in the case of phenol purification) are lowered to the basic values for each DNA sample. The effect of higher Cq value for PCR on native DNA may be a consequence of the influence of neighboring parts of DNA molecules that prevent an amplification or a result of possible interactions of extended DNA molecules with each other. In future work, we plan to study in detail the effect of DNA regions outside the amplicon on RT PCR with a TaqMan probe.
Thus, when performing RT PCR, it is necessary to use DNA preparations obtained by the column purification and digested with a restriction enzyme that cleaves out the amplified fragment. RT PCR of DNA preparations obtained by the phenol method, as well as RT PCR without DNA treatment with a corresponding restriction enzyme, may result in the higher Cq values.
CITATIONS
- Akishev A.G., Netesova N.A., Abdurashitov M.A., Degtyarev S.Kh. Determination of SNP 5mC/T in AluSx repeat (Chr16: 75033884) in DNA samples from the human blood by GlaI- and FatI-PCR assay. Epigenet DNA diagnx, vol.1, 2021, DOI: 10.26213/SE.2021.11.76.001
- Akishev A.G., Netesova N.A., Abdurashitov M.A., Degtyarev S.Kh. Determination of SNP 5mC/T in position Chr1: 245618129 in DNA samples from the human blood by GlaI- and FatI-PCR assay. Epigenet DNA diagnx, vol.1, 2021, p.13-23, DOI: 10.26213/SE.2019.69.42836
- Akishev A.G., Netesova N.A., Abdurashitov M.A., Vihlyanov I.V., Nikitin M.K., Karpov A.B., Degtyarev S.Kh. (2021) Determination of the risk of breast and stomach cancer by identifying variants of the 5mC/T polymorphism in the intron of the KIF26B gene at position xp1: 245618129, Epigenet DNA diagnx, vol.2021(1), DOI: 10.26213/SE.2019.78.40307
- Akishev A.G., Netesova N.A., Abdurashitov M.A., Vihlyanov I.V., Nikitin M.K., Karpov A.B., Degtyarev S.Kh. (2019) GlaI-PCR analysis of methylation of ACGC site in Chr11: 65647266 in DNA samples from the blood cells of healthy donors and early stage breast cancers. Epigenet DNA diagnx, vol. 2019(1), DOI: 10.26213/SE.2019.66.12.001
- EV Dubinin, AG Akishev, MA Abdurashitov, SB Oleynikova, VL Sitko, and S Kh Degtyarev Real time GlaI-PCR assay of regulation regions of human genes HDAC4, RARB and URB1 // Research Journal of Pharmaceutical, Biological and Chemical Sciences, vol 7(2), pp. 667-676 (2016).
- PCR kit with restriction enzymes pretreatment // http://russia.sibenzyme.com/info7873.php
- Biorad CFX96 Instruction manual // https://www.bio-rad.com/webroot/web/pdf/lsr/russian_federation/russian/literature/10021337_ru.pdf
- DNA isolation kit // http://russia.sibenzyme.com/info1561.php