Comparison of the substrate specificity of GlaI recombinant site-specific methyl-directed DNA endonuclease and the native enzyme isolated from Glacial ice bacterium strain
Valery A. Chernukhin, Vladimir S. Dedkov, Danila A. Gonchar, Murat A. Abdurashitov, Alexander G. Akishev, Tatyana N. Nayakshina, Elena N. Lomakovskaya and Sergey Kh. Degtyarev
Valery A. Chernukhin, Vladimir S. Dedkov, Danila A. Gonchar, Murat A. Abdurashitov, Alexander G. Akishev, Tatyana N. Nayakshina, Elena N. Lomakovskaya and Sergey Kh. Degtyarev*
SibEnzyme Ltd., Novosibirsk, 630117, Russia
* the Author for correspondence: E-mail: degt@sibenzyme.com
A cloning of GlaI site-specific methyl-directed DNA endonuclease gene and the comparison of the substrate specificity of recombinant and native enzymes is described. The analysis of recombinant GlaI endonuclease properties showed that the substrate specificity of native and recombinant enzymes didn’t differ, but the concentration of recombinant enzyme exceeded 10 times of native enzyme.
Abbreviations: 5mC – 5-methylcytosine, BSA – bovin serum albumin, DMSO – dimethylsulfoxide, DTT – dithiothreitol, EDTA – ethylenediaminetetraacetic acid, IPTG – Isopropyl-ß-D-1-thiogalactopyranoside, MD endonuclease – methyl-directed DNA endonuclease, MTase – DNA Methyltransferase, N – any nucleotide, PAAG – polyacrylamide gel, Tris – tris-(oxymethyl)-aminomethane
INTRODUCTION
Site-specific methyl-directed DNA endonucleases, or MD endonucleases, are enzymes that recognize and cleave only methylated DNA sequences, without hydrolyzing unmodified DNA [1]. These enzymes are similar to well-studied restriction endonucleases, as they don’t require any cofactors except for Mg2+ ions for their activity. The GlaI MD endonuclease, isolated from the bacterial strain Glacial ice bacterium, recognizes the DNA sequence 5’-R(5mC)GY-3’ (where R is purine and Y is pyrimidine) in the presence of 5-methylcytosine in a complementary chain, and cleaves both chains of DNA in the middle of the site, forming blunt ends [2]. It is noteworthy that mammalian DNMT3 DNA methyltransferase modifies DNA de novo with the formation of the sequence 5’-R(5mC)GY-3’ [3]. Therefore, GlaI has found applications in epigenetic DNA diagnostics, such as successfully determining the methylation status of local sites or fragments of DNA [4,5] and establishing the human epigenome [6].
Since GlaI’s application in epigenetic DNA diagnostics is effective only when complete specific DNA digestion of the analyzed DNA samples is achieved, a high enzyme concentration is necessary. However, a large amount of Glacial ice bacterium strain biomass can only partially solve the problem of enzyme production with high concentration. Cloning the GlaI gene in E.coli allows for a significant increase in enzyme concentration in biomass and the production of highly active recombinant enzyme from a relatively small amount of bacterial cells.
The site-specific methyl-directed DNA endonucleases are new and poorly studied enzymes. In this work, we have undertaken a comparison of the substrate specificity of native and recombinant enzymes.
MATERIALS AND METHODS
Reagents from following manufacturers were used in this study: “Sigma-Aldrich” (USA), “Fisher” (USA), “Panreac” (Spain) and “Helicon” (Russia). The following resins were used for chromatographic purification of enzyme: Phosphocellulose P-11 (“Whatman”, England), Heparin-sepharose (“Bio-Rad”, USA), Hydroxyapatite (“BioRad”, USA). The strain was grown using the medium components made by ”Organotechnie” (France).
The Esherichia coli ER2267 strain (F’ proA B lacIq Δ(lacZ) M15 zzf:: mini-Tn10 (Kanr)/Δ (argF-lacZ)U169 glnV44 e14 (McrA-) rfbD1. recA1 relA1. endA1 spoT1. Thi-1 Δ(mcrC-mrr) 114:: IS10) was from “New England Biolabs, Inc.” (USA). For experiments we used DNA of λ bacteriophage and a variety of C5-methylated plasmids which were constructed before [7-9]. We used 1 kb DNA Ladder (“SibEnzyme”, Russia) as a marker of the molecular weight of DNA fragments. For GlaI enzyme preparation dilution the SE Buffer “B100” (10 mM Tris-HCl, pH 7.6, 50 mM KCl, 0.1 mM EDTA, 200 μg/ml BSA, 1 mM DTT, 50% glycerol) (“Sibenzyme”, Russia) was used. All experiments on the activity assay and substrate specificity determination of recombinant and native GlaI were carried out in 20 μl of the reaction mixture containing the SE Buffer “Y” (33 mM Tris-acetate, pH 7.9, 10 mM Mg-acetate, 66 mM K-acetate, 1 mM DTT) and 0.5 μg of substrate DNA at 30°C for 1 hour.
GlaI gene cloning
The DNA of Glacial ice bacterium strain was isolated as described earlier [10], and it was partially digested with Kzo9I restriction endonuclease (5’-GATC-3’). The Kzo9I-fragments were ligated in pUC19 vector [11] previously linearized by BamHI restriction endonuclease, and, then, the library of the recombinant clones E.coli ER2267 carrying plasmids with Glacial ice bacterium genome DNA fragments was obtained by a standard technique [12]. The clones were analyzed on the presence of GlaI activity in a bacterial cell lysate using pHspAI plasmid DNA [9] previously linearized by DriI restriction endonuclease (pHspAI/DriI) by the method described earlier [13].
Growing cells of E.coli ER2267 (pMTL22-GlaI) recombinant strain carrying the gene of GlaI
For recombinant GlaI biomass obtaining a single colony of the E.coli ER2267 (pMTL22-GlaI) strain was sowed in 0.5-liter flasks with 100 ml of LB medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, рН 7.5) with addition of ampicillin (50 μg/ml). The culture was grown overnight in the thermostat at 37°C. Then the inoculum was sowed to 12 flasks (5 ml inoculum to each flask) with 100 ml of LB medium containing ampicillin. The culture grew in thermoshaker within 16 hours at 30°C with stirring at 140 rpm. After 11 hours of cultivation the IPTG was added into the culture to 1 mM, after that it grew more 5 hours. Then 1 ml of cultural liquid was picked out for the analysis, and the cells were collected by centrifugation for 30 minutes at 3000 g on the Beckman J2-21 centrifuge (Beckman, USA) and were frozen at -20°C.
For the analysis of target activity, the cells from 1 ml of cultural liquid were precipitated by centrifugation on the Eppendorf centrifuge at 12000 rpm for 2 min, and were suspended in 100 μl of the Lysis-buffer (10 mM Tris-HCl, pH 8.5, 0.1 mg/ml Lysozyme, 50 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) for 30 min using “Multi-vortex V-32” (“Biosan”, Lithuania). 8 μl of crude lysate was added to 40 μl of the reaction mixture containing 1x SE buffer “Y” and 0.5 μg of pHspAI/DriI plasmid DNA which was used earlier for native GlaI activity assay [9] in the microplate well. The suspension was mixed by pipetting and 20 μl of this one was transferred to the following well with 20 μl of the same reaction mixture to make the dilution of the lysate in 2 time. Such procedure was repeated to 7 passage. Reaction mixture was incubated for 1 hour at 30°C, and the reaction was stopped by an addition of the Stop-buffer (40% sucrose, 0.1 M EDTA, 0.05% bromophenol blue).
Isolation of recombionant GlaI enzyme from biomass
All procedures of enzyme isolation were performed at 4°C. 40 g of frozen cells of E.coli ER2267 (pMTL22-GlaI) were suspended in 30 ml of Buffer A (10 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 7 mM β-mercaptoethanol) containing 0.3 M NaCl, 0.3 mg/ml Lysozyme and 0.1 mM phenylmethylsulphonyl fluoride (PMSF). Biomass was lysed in a glass flask with stirring on a magnetic stirrer for 1 hour. As a result of cells lysis the solution became more viscous and transparent. The obtained lysate was disrupted by ultrasonic disintegrator Soniprep 150 (MSE, England) with an adapter diameter of 2 cm, 5 times of 30 seconds with intervals in 1 min for cooling of suspension in an ice bath. Further the lysate was clarified by centrifugation on J2-21 centrifuge (“Beckman”, USA) at 15000 rpm for 30 min.
The supernatant was initially applied to a 20 ml of Phosphocellulose P-11 column pre-equilibrated with Buffer A containing 0.1 M NaCl, and washed with two column volumes of the same buffer. Enzyme elution was performed with 200 ml of a linear gradient of NaCl concentration (0.2-0.65 M) in Buffer A. 20 fractions of 10 ml were collected. Endonuclease-containing fractions were pooled and dialyzed against 20 volumes of Buffer A with 0.1 M NaCl and applied on a column with 10 ml of Heparin-sepharose. Enzyme elution was carried out with 200 ml of a linear gradient of NaCl (0.1-0.45 M) in Buffer A. 50 fractions of 4 ml were collected. Target fractions were pooled and applied on the column with 4 ml of Hydroxyapatite pre-equilibrated with Buffer B (10 mM K-phosphate, pH 7.2, 0.1 mM EDTA, 7 mM β-mercaptoethanol) containing 0.02 M NaCl. The column was washed with 10 ml of the same buffer, and target protein was eluted by a linear gradient of K-phosphate buffer, pH 7.2, from 0.01 to 0.4 M K-phosphate with a volume of 100 ml. 40 fractions of 2.5 ml were collected. Active fractions were pooled, dialyzed against 20 volumes of the concentrating buffer (10 mM Tris-HCl, pH 7.5, 0.1 M NaCl, 0.1 mM EDTA, 0.05% Triton X-100, 7 mM β-mercaptoethanol, 50% glycerol). The enzyme preparation was stored at -20°C.
RESULTS AND DISCUSSION
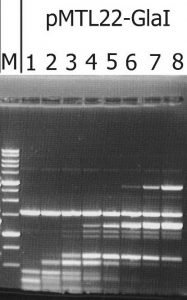
The recombinant plasmid with the target gene of GlaI was isolated as a result of determining the GlaI MD endonuclease activity in lysates of E.coli clones from the pUC19/BamHI-Gla DNA/Kzo9I library, using the pHspAI/DriI DNA substrate. Subsequently, the PCR fragment, including the target gene, was ligated into the FauNDI-BamHI sites of pMTL22 [14]. E.coli ER2267 cells were transformed using the resulting construct, pMTL22-GlaI, and the recombinant strain, E.coli ER2267 (pMTL22-GlaI), was selected and used for biomass growth and subsequent enzyme isolation. The results of testing the target activity in a lysate of cells from the recombinant E.coli ER2267 (pMTL22-GlaI) biomass are shown in Figure 1.
Figure 1. The target activity analysis in biomass of the recombinant strain-producer – E.coli ER2267 (pMTL22-GlaI), on pHspAI/DriI plasmid DNA.
Lanes: 1-8 – serial twice lysate dilution of the biomass, since 4 μl of lysate; M – 1 kb molecular weight DNA ladder (from 0,25 to 10 kb). Electrophoresis in 1% agarose gel.
As shown in Figure 1, complete hydrolysis of the substrate DNA was observed in lane 1, and the resulting fragments set was typical of native GlaI [9].
Chromatographic purification resulted in the isolation of 6 ml of enzyme preparation with an activity of 100,000 units/ml from 10 g of the recombinant strain-producer. This was ten times higher than the activity of GlaI isolated from the native strain (up to 10,000 units/ml).
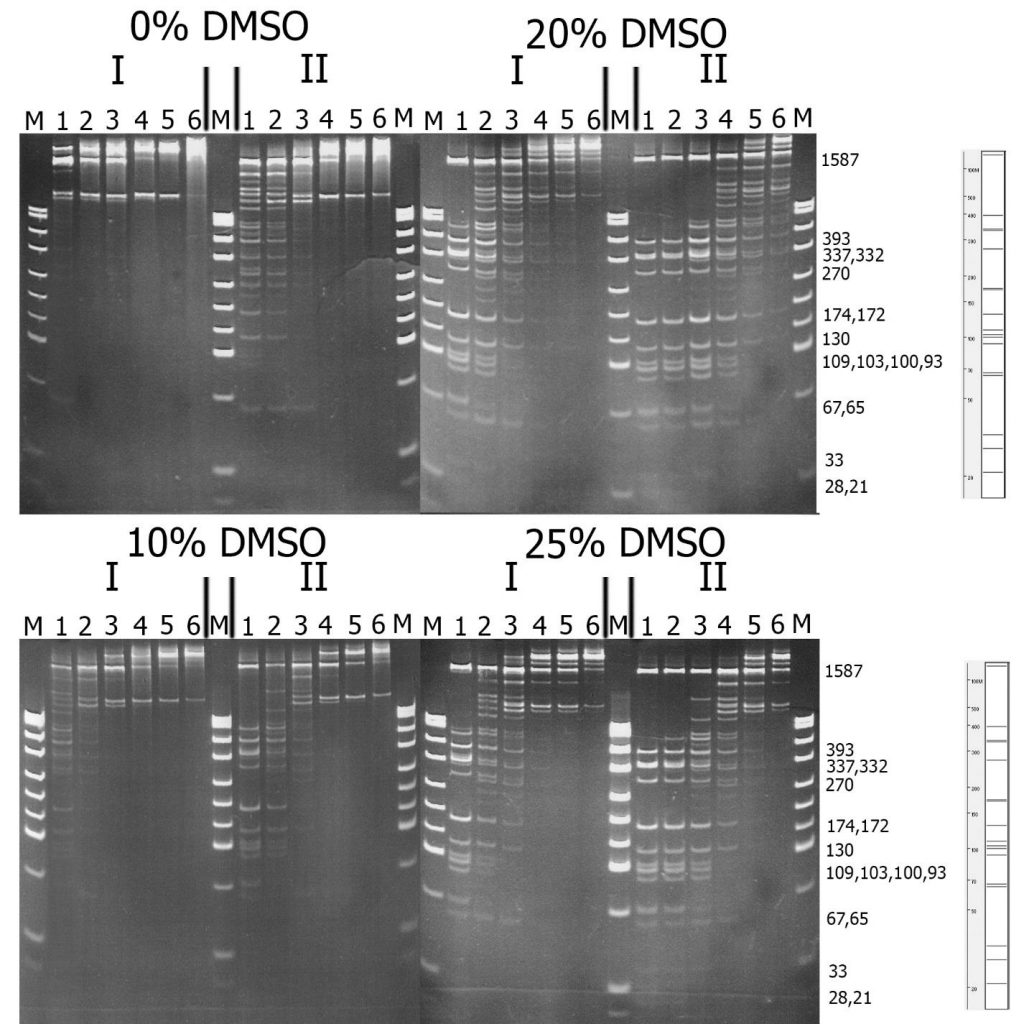
To compare the substrate specificity of recombinant and native GlaI, digestion reactions were carried out on pHspAI/DriI DNA in the presence of different concentrations of DMSO. As previously reported, the activity of native GlaI increases 20-30 times in the presence of 20% DMSO [15]. The reactions were carried out in SE buffer “Y” at the optimal temperature (30°C) for 1 hour.
Figure 2 shows a comparison of the activities of GlaI preparations isolated from native and recombinant strains. For ease of comparison, the recombinant GlaI preparation was diluted four times with SE buffer for enzyme storage and dilution “B100”.
Figure 2. Comparison of substrate specificity of GlaI isolated from the native (I) and recombinant (II) bacterial strains on pHspAI/DriI DNA. Electrophoresis in 12% PAAG. On the right theoretically calculated picture of pHspAI/DriI plasmid digestion on the sites 5’-R(5mC)GY-3’ is given.
Lanes: 1-6 – serial twice enzyme preparation dilution, since 1 μl of the preparations; M – pUC19/MspI molecular weight DNA ladder.
From Figure 2, it is evident that recombinant GlaI, like the native enzyme, is activated in the reaction mixture with DMSO, and the highest activity is observed at a DMSO concentration of 20%. This figure also indicates that the substrate specificity of both enzyme preparations on pHspAI/DriI does not differ significantly. However, DNA digested with the recombinant GlaI preparation, even when diluted by 4 times, displayed more complete digestion of the DNA substrate compared to the native GlaI preparation.
Furthermore, we conducted a comparative analysis of the activities of native and recombinant GlaI preparations on various methylated DNA substrates. We used phage λ DNA and the following plasmid DNA as substrates:
- pMHpaII (which includes the gene of HpaII DNA MTase modifying the second cytosine in the 5 ‘-CCGG-3’ sequence and containing the 5’-C(5mC)GG-3’/3’-GG(5mC)C-5’ sites [16]);
- pMHaeIII (which includes the gene of HaeIII DNA MTase modifying the first cytosine in the 5 ‘-GGCC-3′ sequence and containing the 5’-GG(5mC)C-3’/3’-C(5mC)GG-5’ sites [17]);
- pFsp4HI3 (which includes the gene of Fsp4HI DNA MTase modifying the first cytosine in the 5′-GCNGC-3′ sequence and containing the 5’-G(5mC)NGC-3’/3’-CGN(5mC)G-5’ sites [8]);
- pBspACI (which includes the genes of BspACI-1 and BspACI-2 DNA MTases modifying the cytosines in the 5′-CCGC-3′ and 5′-GCGG-3′ sequences and containing the 5’-(5mC)CGC-3’/3’-GG(5mC)G-5’ sites [18]);
- pHspAI (which includes the gene of HspAI DNA MTase modifying the first cytosine in the 5′-GCGC-3′ sequence and containing the 5’-G(5mC)GC-3’/3’-CG(5mC)G-5’ sites [9]).
For convenient analysis of DNA hydrolysis, all plasmid DNAs were linearized using DriI restriction endonuclease before the reactions. Reactions were performed in SE Buffer “Y” in the presence of 20% DMSO.
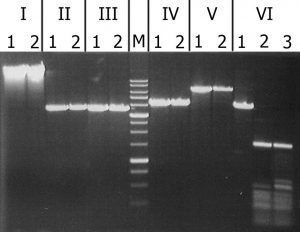
The results of cleavage of these substrates with recombinant GlaI are presented in Figure 3.
igure 3. Analysis of substrate specificity of recombinant GlaI. Electrophoresis in 1% agarose gel. Lanes: I – Lambda; II – pMHpaII/DriI; III – pMHaeIII/DriI; IV-pFspAI3/DriI; V – pBspACI/DriI; VI – pHspAI/DriI; 1 – DNA without enzyme addition; 2 – DNA with addition of recombinant GlaI; 3 – DNA with addition of native GlaI; M – 1 kb molecular weight DNA ladder (from 0,25 to 10 kb).
As can be seen from this figure, recombinant GlaI, like the native enzyme, does not cleave unmethylated DNA substrates (Lambda) and all C5-methylated substrates, except the pHspAI plasmid. This fact confirms that recombinant GlaI, as well as the native one, recognizes and cuts only the sequence 5’-R(5mC)GY-3’ containing the 5-methylcytosine.
The results obtained confirm that the GlaI isolated from the recombinant strain, with 10 times greater activity, and exhibiting the same substrate specificity as the enzyme from the native (Glacial ice bacterium) strain. Therefore, it is possible to conclude that the use of recombinant GlaI allows for more complete digestion of DNA samples, which will undoubtedly improve the results of epigenetic research using this enzyme.
REFERENCES
- http://sibenzyme.com/products/m2_type
- Tarasova G. V., Nayakshina T. N., Degtyarev S. Kh. Substrate specificity of new methyl-directed DNA endonuclease GlaI . // BMC Molecular Biology 2008, 9:7.
- Handa V., Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. // J. Mol. Biol. 2005. 348 (5): 1103-1112.
- Gonchar D.A., Akishev A.G., Degtyarev S.Kh. BlsI- and GlaI-PCR assays – a new method of DNA methylation study. // Ovchinnikov bulletin of biotechnology and physical and chemical biology. 2010. V.6, № 1, P. 5-12.
- Kuznetsov V.V., Abdurashitov M.A., Akishev A.G., Degtyarev S.H. Method of determining nucleotide sequence Pu(5mC)GPy at predetermined position of long-distance DNA. // Russian Federation patent RU 2525710 C1. 2014. (In Russian).
- Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Kuznetsov V.V., Degtyarev S.Kh. Mapping of R(5mC)GY Sites in the Genome of Human Malignant Cell Line Raji. // Biol. Med. (Aligarh). 2015. V. 7. Is. 4. BM-135-15.
- Chmuzh E.V., Kashirina J.G., Tomilova J.E., Mezentseva N.V., Dedkov V.S., Gonchar D.A., Abdurashitov M.A., Degtyarev S.Kh. A Novel Restriction Endonuclease BisI from Bacillus subtilis T30, Recognizes a Methylated DNA Sequence 5′-G(m5C)^NGC-3′. // Biotekhnologia (Moscow). 2005. №.3. P.22-26. (In Russian).
- Chernukhin V.A., Chmuzh E.V., Tomilova Yu.E., Nayakshina T.N., Gonchar D.A., Dedkov V.S., Degtyarev S.Kh. A novel site-specific endonuclease GluI recognizes methylated DNA sequence 5’-G(5mC)^NG(5mC)-3’/3’-(5mC)GN^(5mC)G. // Bulletin of biotechnology and physico-chemical biology named by Yu.A.Ovchinnikov (Moscow). 2007. V.3, №.2, P.13-17. (In Russian).
- Chernukhin V.A., Najakshina T.N., Abdurashitov M.A., Tomilova J.E., Mezentzeva N.V., Dedkov V.S., Mikhnenkova N.A., Gonchar D.A., Degtyarev S. Kh A novel restriction endonuclease GlaI recognizes methylated sequence 5′-G(5mC)^GC-3′. // Biotechnologia. 2006. V 4. P.31-35. (In Russian).
- Smith C.L., Klco S.R. and Cantor C.R. In: Davies K. (ed.). Genome Analysis: A Practical Approach. 1987. IRL Press. Oxford. UK.